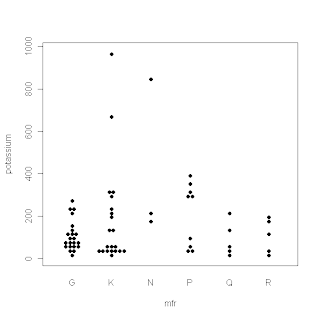
My last four posts have dealt with boxplots and some useful variations on that theme. Just after I finished the series, Tal Galili, who maintains the R-bloggers website, pointed me to a variant I hadn’t seen before. It's called a beeswarm plot, and it's produced by the beeswarm package in R. I haven’t played with this package a lot yet, but it does appear to be useful for datasets that aren’t too large and that you want to examine across a moderate number of different segments. The plot shown below provides a typical illustration: it shows the beeswarm plot comparing the potassium content of different cereals, broken down by manufacturer, from the UScereal dataset included in the MASS package in R. I discussed this data example in my first couple of boxplot posts and I think this is a case where the beeswarm plot gives you a more useful picture of how the data points are distributed than the boxplots do. For more information about the beeswarm package, I recommend Tal's post. More generally, anyone interested in learning more about what you can do with the R software package should find the R-blogger website extremely useful.
Besides boxplots, one of the other useful graphical data characterizations I discuss in Exploring Data in Engineering, the Sciences, and Medicine is the quantile-quantile (Q-Q) plot. The most common form of this characterization is the normal Q-Q plot, which represents an informal graphical test of the hypothesis that a data sequence is normally distributed. That is, if the points on a normal Q-Q plot are reasonably well approximated by a straight line, the popular Gaussian data hypothesis is plausible, while marked deviations from linearity provide evidence against this hypothesis. The utility of normal Q-Q plots goes well beyond this informal hypothesis test, however, which is the main point of this post. In particular, the shape of a normal Q-Q plot can be extremely useful in highlighting distributional asymmetry, heavy tails, outliers, multi-modality, or other data anomalies. The specific objective of this post is to illustrate some of these ideas, expanding on the discussion presented in Exploring Data.
is the quantile-quantile (Q-Q) plot. The most common form of this characterization is the normal Q-Q plot, which represents an informal graphical test of the hypothesis that a data sequence is normally distributed. That is, if the points on a normal Q-Q plot are reasonably well approximated by a straight line, the popular Gaussian data hypothesis is plausible, while marked deviations from linearity provide evidence against this hypothesis. The utility of normal Q-Q plots goes well beyond this informal hypothesis test, however, which is the main point of this post. In particular, the shape of a normal Q-Q plot can be extremely useful in highlighting distributional asymmetry, heavy tails, outliers, multi-modality, or other data anomalies. The specific objective of this post is to illustrate some of these ideas, expanding on the discussion presented in Exploring Data.
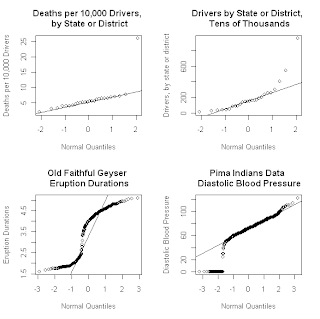
The above figure shows four different normal Q-Q plots that illustrate some of the different data characteristics these plots can emphasize. The upper left plot demonstrates that normal Q-Q plots can be extremely effective in highlighting glaring outliers in a data sequence. This plot shows the annual number of traffic deaths per ten thousand drivers over an unspecified time period, for 25 of the 50 states in the U.S. , plus the District of Columbia Maine
It is not clear why the reported traffic death rate is so high for Maine Maine
The Q-Q plot for this denominator variable – i.e., for the number of drivers – is shown as the upper right plot in the original set of four shown above. There, the fact that both tails of the distribution lie above the reference line is suggestive of distributional asymmetry, a point examined further below using Q-Q plots for other reference distributions. Also, note that both of the upper Q-Q plots shown above are based on only 26 data values, which is right at the lower limit on sample size that various authors have suggested for normal Q-Q plots to be useful (see the discussion of normal Q-Q plots in Section 6.3.3 of Exploring Data for details). The tricky issues of separating outliers, asymmetry, and other potentially interesting data characteristics in samples this small is greatly facilitated using the Q-Q plot confidence intervals discussed below.
The lower left Q-Q plot in the above sequence is that for the Old Faithful geyser dataset faithful included with the base R package. As I have discussed previously, the eruption duration data exhibits a pronounced bimodal distribution, which may be seen clearly in nonparametric density estimates computed from these data values. Normal Q-Q plots constructed from bimodal data typically exhibit a “kink” like the one seen in this plot. A crude way of explaining this behavior is the following: the lower portion of the Q-Q plot is very roughly linear, suggesting a very approximate Gaussian distribution, corresponding to the first mode of the eruption data distribution (i.e., the durations of the shorter group of eruptions). Similarly, the upper portion of the Q-Q plot is again very roughly linear, but with a much different intercept that corresponds to the larger mean of the second peak in the distribution (i.e., the durations of the longer group of eruptions). To connect these two “roughly linear” local segments, the curve must exhibit a “kink” or rapid transition region between them. By the same reasoning, more general multi-modal distributions will exhibit more than one such “kink” in their Q-Q plots. Finally, the lower right Q-Q plot in the collection above was constructed from the Pima Indians diabetes dataset available from the UCI Machine Learning Repository. This dataset includes a number of clinical measurements for 768 female members of the Pima tribe of Native Americans, including their diastolic blood pressure. The lower right Q-Q plot was constructed from this blood pressure data, and its most obvious feature is the prominent lower tail anomaly. In fact, careful examination of this plot reveals that these points correspond to the value zero, which is not realistic for any living person. What has happened here is that zero has been used to code missing values, both for this variable and several others in this dataset. This observation is important because the metadata associated with this dataset indicates that there is no missing data, and a number of studies in the classification literature have proceeded under the assumption that this is true. Unfortunately, this assumption can lead to badly biased results, a point discussed in detail in a paper I published in SIGKDD Explorations (Disguised Missing Data paper PDF). The point of the example presented here is to show that normal Q-Q plots can be extremely effective in highlighting this kind of data anomaly.
The normal Q-Q plots considered so far were constructed using the qqnorm procedure available in base R, and the reference lines shown in these plots were constructed using the qqline command. It is not difficult to construct Q-Q plots for other reference distributions using procedures in base R, but a much simpler alternative is to use the qqPlot command in the optional car package. This R add-on package was developed in association with the book An R Companion to Applied Regression , by Fox and Weisberg, and it includes a number of very useful procedures. The default options of the qqPot procedure automatically generate a reference line, along with upper and lower 95% confidence intervals for the plot, which are particularly useful for small samples like the road dataset. The figure below shows a normal Q-Q plot for the number of traffic deaths per 10,000 drivers generated using the qqPlot package. The fact that all of the points but the one obvious outlier fall within the 95% confidence limits suggest that the scatter around the reference line seen for these 25 observations is small enough to be consistent with a normal reference distribution. Further, these confidence limits also emphasize how much the outlying result for the state of
, by Fox and Weisberg, and it includes a number of very useful procedures. The default options of the qqPot procedure automatically generate a reference line, along with upper and lower 95% confidence intervals for the plot, which are particularly useful for small samples like the road dataset. The figure below shows a normal Q-Q plot for the number of traffic deaths per 10,000 drivers generated using the qqPlot package. The fact that all of the points but the one obvious outlier fall within the 95% confidence limits suggest that the scatter around the reference line seen for these 25 observations is small enough to be consistent with a normal reference distribution. Further, these confidence limits also emphasize how much the outlying result for the state of Maine
Another advantage of the qqPlot command is that it provides the basis for very easy generation of Q-Q plots for essentially any reference distribution that is available in R, including those available in add-on packages like gamlss.dist, which supports an extremely wide range of distributions (generalized inverse Gaussian distributions, anyone?). This capability is illustrated in the four Q-Q plots shown below, all generated with the qqPlot command for non-Gaussian distributions. In all of these plots, the data corresponds to the driver counts for the 26 states and districts summarized in the road dataset. Motivation for the specific Q-Q plots shown here is that the four distributions represented by these plots are all better suited to capturing the asymmetry seen in the normal Q-Q plot for this data sequence than the symmetric Gaussian distribution is. The upper left plot shows the results obtained for the exponential distribution which, like the Gaussian distribution, does not require the specification of a shape parameter. Comparing this plot with the normal Q-Q plot shown above for this data sequence, it is clear that the exponential distribution is more consistent with the driver data than the Gaussian distribution is. The data point in the extreme upper right does fall just barely outside the 95% confidence limits shown on this plot, and careful inspection reveals that the points in the lower left fall slightly below these confidence limits, which become quite narrow at this end of the plot.
The exponential distribution represents a special case of the gamma distribution, with a shape parameter equal to 1. In fact, the exponential distribution exhibits a J-shaped density, decaying from a maximum value at zero, and it corresponds to a “dividing line” within the gamma family: members with shape parameters larger than 1 exhibit unimodal densities with a single maximum at some positive value, while gamma distributions with shape parameters less than 1 are J-shaped like the exponential distribution. To construct Q-Q plots for general members of the gamma family, it is necessary to specify a particular value for this shape parameter, and the other three Q-Q plots shown above have done this using the qqPlot command. Comparing these plots, it appears that increasing the shape parameter causes the points in the upper tail to fall farther outside the 95% confidence limits, while decreasing the shape parameter better accommodates these upper tail points. Conversely, decreasing the shape parameter causes the cluster of points in the lower tail to fall farther outside the confidence limits. It is not obvious that any of the plots shown here suggest a better fit than the exponential distribution, but the point of this example was to show the flexibility of the qqPlot procedure in being able to pose the question and examine the results graphically. Alternatively, the Weibull distribution – which also includes the exponential distribution as a special case – might describe these data values better than any member of the gamma distribution family, and these plots can also be easily generated using the qqPlot command (just specify dist = “weibull” instead of dist = “gamma”, along with shape = a for some positive value of a other than 1).
Finally, one cautionary note is important here for those working with very large datasets. Q-Q plots are based on sorting data, something that can be done quite efficiently, but which can still take a very long time for a really huge dataset. As a consequence, while you can attempt to construct Q-Q plots for sequences of hundreds of thousands of points or more, you may have to wait a long time to get your plot. Further, it is often true that plots made up of a very large number of points reduce to ugly-looking dark blobs that can use up a lot of toner if you make the further mistake of trying to print them. So, if you are working with really enormous datasets, my suggestion is to construct Q-Q plots from a representative random sample of a few hundred or a few thousand points, not hundreds of thousands or millions of points. It will make your life a lot easier.